 |
Blue Star STING Quick Learn |
 |
Blue Star STING Quick Learn |
We would like to be as silent as possible here!
Pictures are going to be your guide and just here and there we will give some
textual instructions.
One picture is worth 1000 words. And we hope that pictures are descriptive;
Enjoy the cruise! And, by the way, if you wish to be faster in Quick_Learning,
try DIMOND STING WARP Speed Results!
See also this application! And see how to
obtain images as in our
publication published by NAR Special Web server issue (2003).
We highly recommend that the user gets acquainted with information that PDB
files are carrying; use PDB_Metrics
for this purpose!
| Basic commands | Modules |
| STING Millennium Modes | Data Parsing --- Blue Star STING: Links |
|
|
 |
STING Millennium Quick LearnBasic commands |
|
To make right choice of your browser,
please see here!
|
| To see the 3D structure you need to download and install the CHIME plugin |
Also, chances are very small, but you may be interested in one of "very specific PDB files" and there will be very specific STING response! There are also "very creative residue numbering in PDB files". See about known STING bugs here! Also, get acquainted with general characteristics of PDB through our PDB_Metrics!
Let us analyze 1dhk.pdb (alpha amylase in complex with inhibitor):
| Left Image: |
|
Right Image:
|
Please learn more about STING Millennium basic
features!
We hope that no words are necessary here! To get from the display at the upper
right image to the lower left image, just follow magenta
line instructions!
| Left Image: |
Ribbons !
Note: water molecule positions are indicated by red wireframe presentation.
Note #2: Try here experimenting and see
what type of img you will get by successively clicking
on the REFRESH button!
| Right Image: |
See more of ACTION menu options!
|
|
| Left Image: |
| Right Image: |
|
|
See more options on Interface
On menu!
See more options on Int Surface menu!
| Left Image: |
|
Right Image:
|
| Left Image: |
|
Image bellow:
|
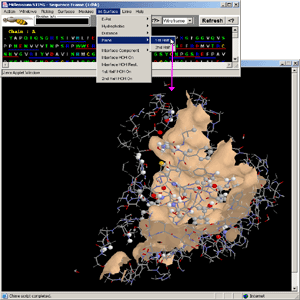
In this example, the user can observe just the "1st half"
of the interface as a solid surface. This is the part of the interface designated
at "Activate menu Options - Right Image" as
chain A. The spacefill presentation is done only for the atoms that are within
contact distance from the atoms belonging to the Interface Forming
Residues (IFR) from the opposite side of the presented
surface. All IFRs are displayed in wireframe presentation.
|
Go to Quick Learn: Modules
|
Go to Quick Learn: Modes
|