ROTAMER
We have used the Rotamer Library presented in Lovell et. Al. [1]. This library can be obtained at ftp://kinemage.biochem.duke.edu/pub/datasets/rotamer/rotamerDocs/Win/Complete_rotamer_lib.txt
All ![]() torsion
angles are calculated according to the definition found in “A to
Z of O” (http://xray.bmc.uu.se/~alwyn/A-Z_of_O/A-Z_frameset.html).
torsion
angles are calculated according to the definition found in “A to
Z of O” (http://xray.bmc.uu.se/~alwyn/A-Z_of_O/A-Z_frameset.html).
Placing the cursor above this element: pop-up area will
show the position (sequence number and AA three letter code) for selected
amino acid, the rotamer’s name according to [1], percentage and
the torsion angles of the selected amino acid.

The "Rotamer" color coding represents the rotamer percentage
of occurrence. There are two color coding schemes:
- First row: from 0 to 1, where 0 is 0% (dark blue) of occurrence and 1, 100% (blue).
- Second row: from the minimum to the maximum percentage of occurrence of each amino acid.
- The last row indicates
in yellow the amino acids that do not have
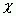 torsion angles (GLY, ALA and PRO), and in red the rare rotamers. Rare
rotamers are defined as those torsion angles that are not close enough
(Euclidean distance greater than a given limit. This limit is calculated
using the assumption that each torsion angle
torsion angles (GLY, ALA and PRO), and in red the rare rotamers. Rare
rotamers are defined as those torsion angles that are not close enough
(Euclidean distance greater than a given limit. This limit is calculated
using the assumption that each torsion angle 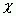 goes from
goes from 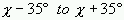 ) to any
one of the rotamers of an amino acid or has the probability of occurrence
less then 1%.
) to any
one of the rotamers of an amino acid or has the probability of occurrence
less then 1%.
Left mouse click:
no action
Right mouse click: on any of the "Rotamer" will generate following
menu and actions:
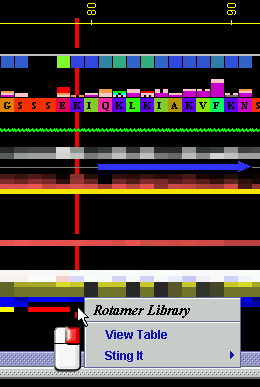
A) View Table
“View Table” will produce the table:
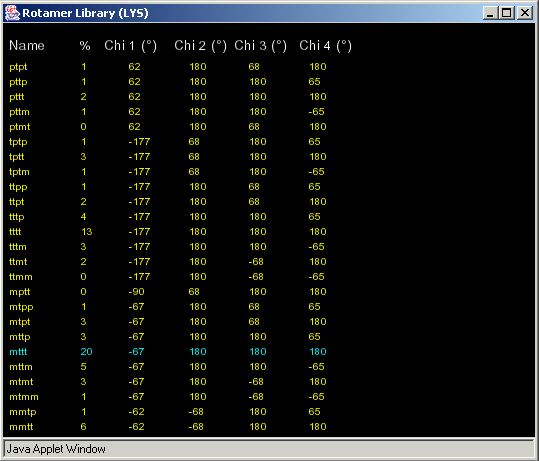
In a different color is the rotamer that is near to the torsion angles of the selected amino acid.
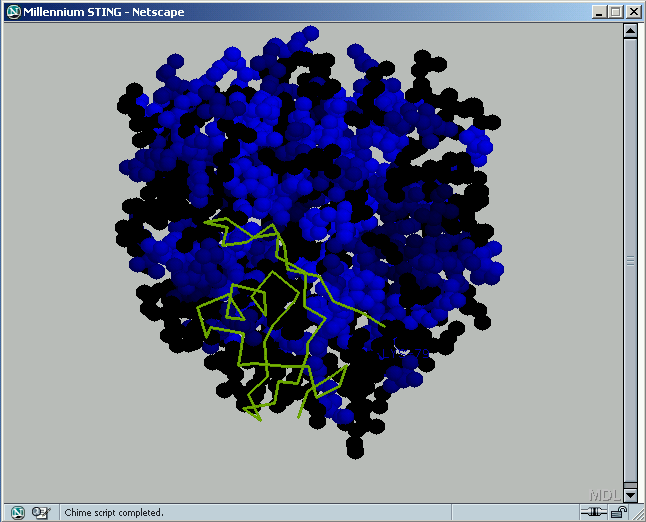
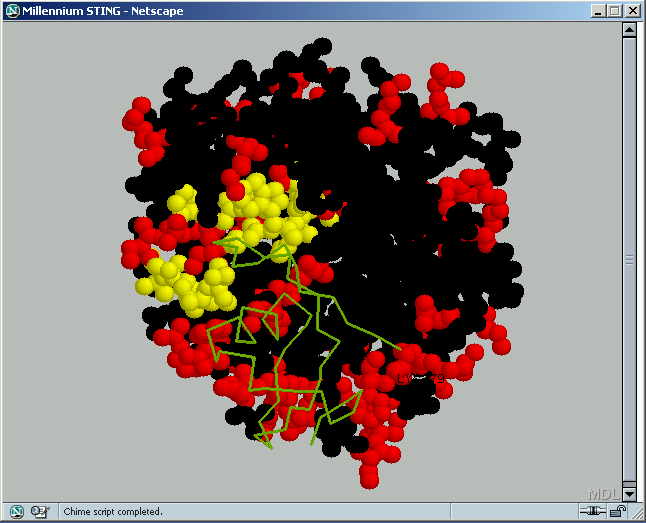
B) STING it : This option generates structural presentation with color coding of the amino acids corresponding to the "Rotamers" color coding. Amino acids are presented in CPK rendering.
B1) %
B2) Rare Rotamers
[1] Lovell,S. C., Word, J. M., Richardson, J. S. and Richardson, D. C., (2000) “The Penultimate Rotamer Library”, PROTEINS: Structure, Function and Genetics, 40:389-408