ProTherm
ProTherm is a collection of numerical data of thermodynamic parameters such as Gibbs free energy change, enthalpy change, heat capacity change, transition temperature etc. for wild type and mutant proteins, that are important for understanding the structure and stability of proteins. It also contains information about secondary structure and accessibility of wild type residues, experimental conditions (pH, temperature, buffer, ion and protein concentration), measurements and methods used for each data, and activity information (Km and Kcat ).
We have taken the ProTherm values from the Thermodynamic Database for Proteins and Mutants.
We currently have all ProTherm corss-links for a "wild type proteins" (non-mutated ones) in a following PDB structures:(click here to see the list)
Placing the cursor above this element: pop-up area will show the position
(sequence number and AA three letter code) for selected amino acid and
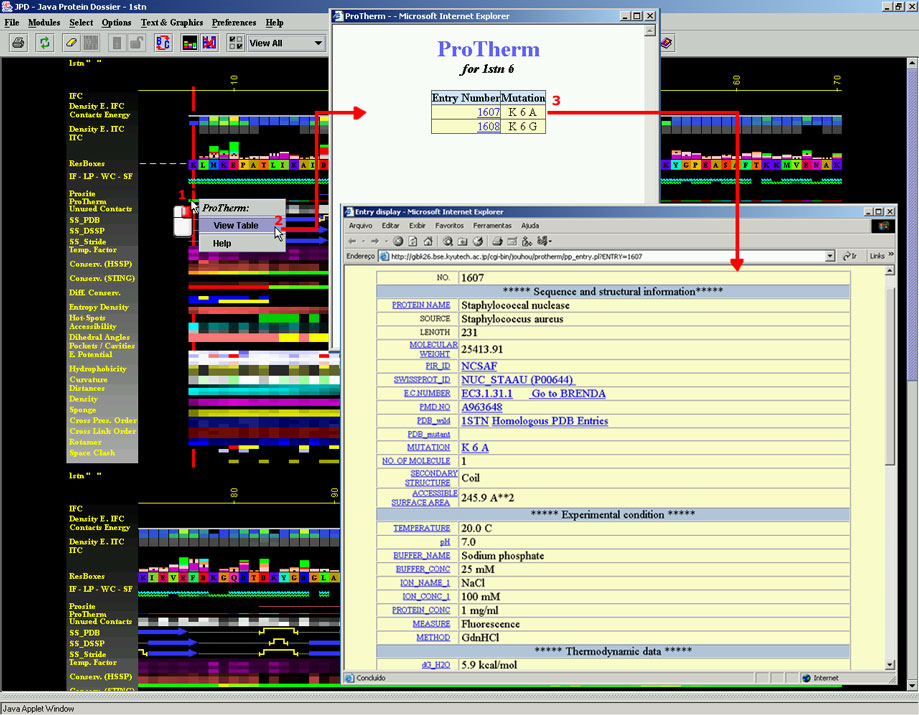
the pop up menu will offer to a user to display the ProTherm Table. This
table contains one or more pointers, indicating ProTherm ID and the type
of mutation. In the figure 1. below, we can see that for the protein 1stn.pdb,
position 15 which is originally occupied by Isoleucine, is mutated to
V (ProTherm ID 459), to A (ProTherm ID 460) etc. A user can then opt to
see details annotated in ProTherm for any of those mutations. The Figure
2. below shows a typical ProTherm page with information on specific mutation.
Left mouse click: no action
Right mouse click: on any of the "ProTherm elements" will generate
following menu and actions:
Figure 1.
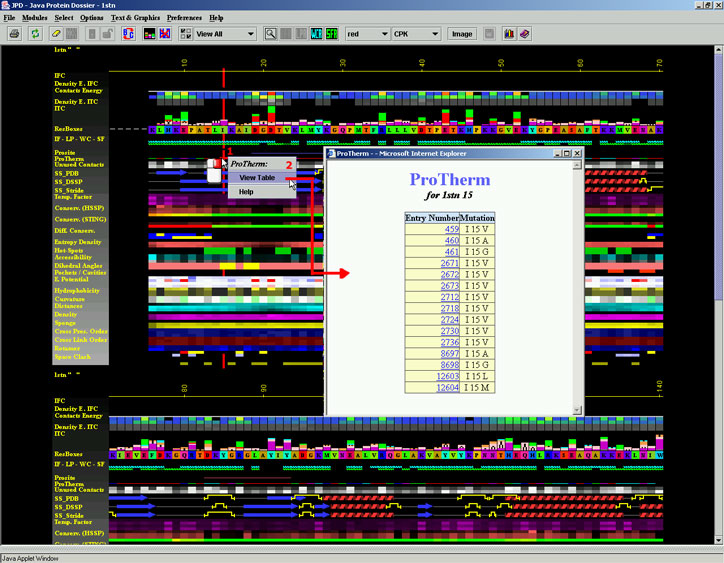
Figure 2.